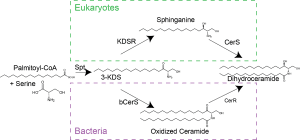
Bacterial sphingolipid synthesis
Caulobacter crescentus is a Gram-negative oligotrophic organism that is well adapted to life in low phosphate environment. 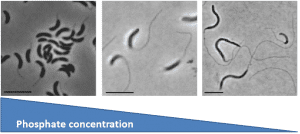 In response to phosphate limitation, the cells and its stalk appendage elongate dramatically leading to an overall increase in cellular surface area of 6-7 fold. Since most membranes, including C. crescentus, are composed of phosphoplipids, how do you make all of this new membrane material in the absence of phosphate? Working with Ziqiang Guan (Duke University), Dominic Campopiano (University of Edinburgh), and Howard Goldfine (University of Pennsylvania), we found that C. crescentus can make glycosphingolipids in response to phosphate starvation. These ceramide-based lipids were thought to be fairly unusual in bacteria, though they are ubiquitous among eukaryotes. In C. crescentus, these lipids play a role in resistance to phage by reducing their adsorption to the cell surface.
In response to phosphate limitation, the cells and its stalk appendage elongate dramatically leading to an overall increase in cellular surface area of 6-7 fold. Since most membranes, including C. crescentus, are composed of phosphoplipids, how do you make all of this new membrane material in the absence of phosphate? Working with Ziqiang Guan (Duke University), Dominic Campopiano (University of Edinburgh), and Howard Goldfine (University of Pennsylvania), we found that C. crescentus can make glycosphingolipids in response to phosphate starvation. These ceramide-based lipids were thought to be fairly unusual in bacteria, though they are ubiquitous among eukaryotes. In C. crescentus, these lipids play a role in resistance to phage by reducing their adsorption to the cell surface. 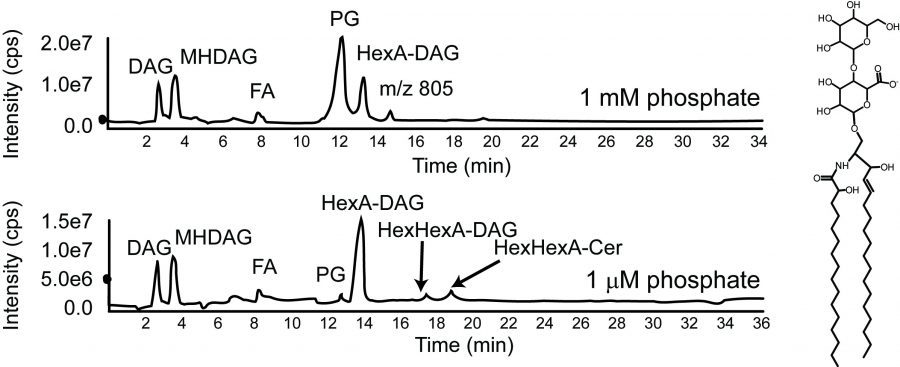 A major reason we thought these lipids are rare in bacteria was that we didn’t understand how these lipids are made in bacteria. While the eukaryotic pathway is well characterized, most of the eukaryotic biosynthesis enzymes do not have bacterial homologues?? We recently discovered that the bacterial synthetic mechanism evolved independently of the eukaryotic pathway and that the chemical reactions occur in a different order! Additionally, it now appears that a huge variety of bacteria encode the genes required for sphingolipid synthesis. Our findings provide many new opportunities to study the physiological roles of the diverse set of bacterial sphingolipids.
A major reason we thought these lipids are rare in bacteria was that we didn’t understand how these lipids are made in bacteria. While the eukaryotic pathway is well characterized, most of the eukaryotic biosynthesis enzymes do not have bacterial homologues?? We recently discovered that the bacterial synthetic mechanism evolved independently of the eukaryotic pathway and that the chemical reactions occur in a different order! Additionally, it now appears that a huge variety of bacteria encode the genes required for sphingolipid synthesis. Our findings provide many new opportunities to study the physiological roles of the diverse set of bacterial sphingolipids.
See more about bacterial sphingolipids in this Webinar given by Eric for the Sphingolipid Biology series